# Covariance matrix that combines random effects and residual (V = ZGZ^t + R)
getV <- function(model) {
Lamda<-getME(model,"Lambda") ##matrix with scaling factor for random sd (relative to sigma)
var.lamda <- t(Lamda)%*%Lamda ## scale for variance (sigma^2)
Z <- getME(model,"Z")
sigma2 <- sigma(model)^2 ##
var.g<-sigma2*var.lamda
var.random <- (Z %*% var.g %*%t(Z))
var.resid <- sigma2 *Diagonal(nrow(Z))
var.y <- var.random + var.resid
invisible(var.y)
}6 Linear mixed models III
6.1 Auxiliary functions
We will use the following function to extract the covariance matrix of a linear mixed model:
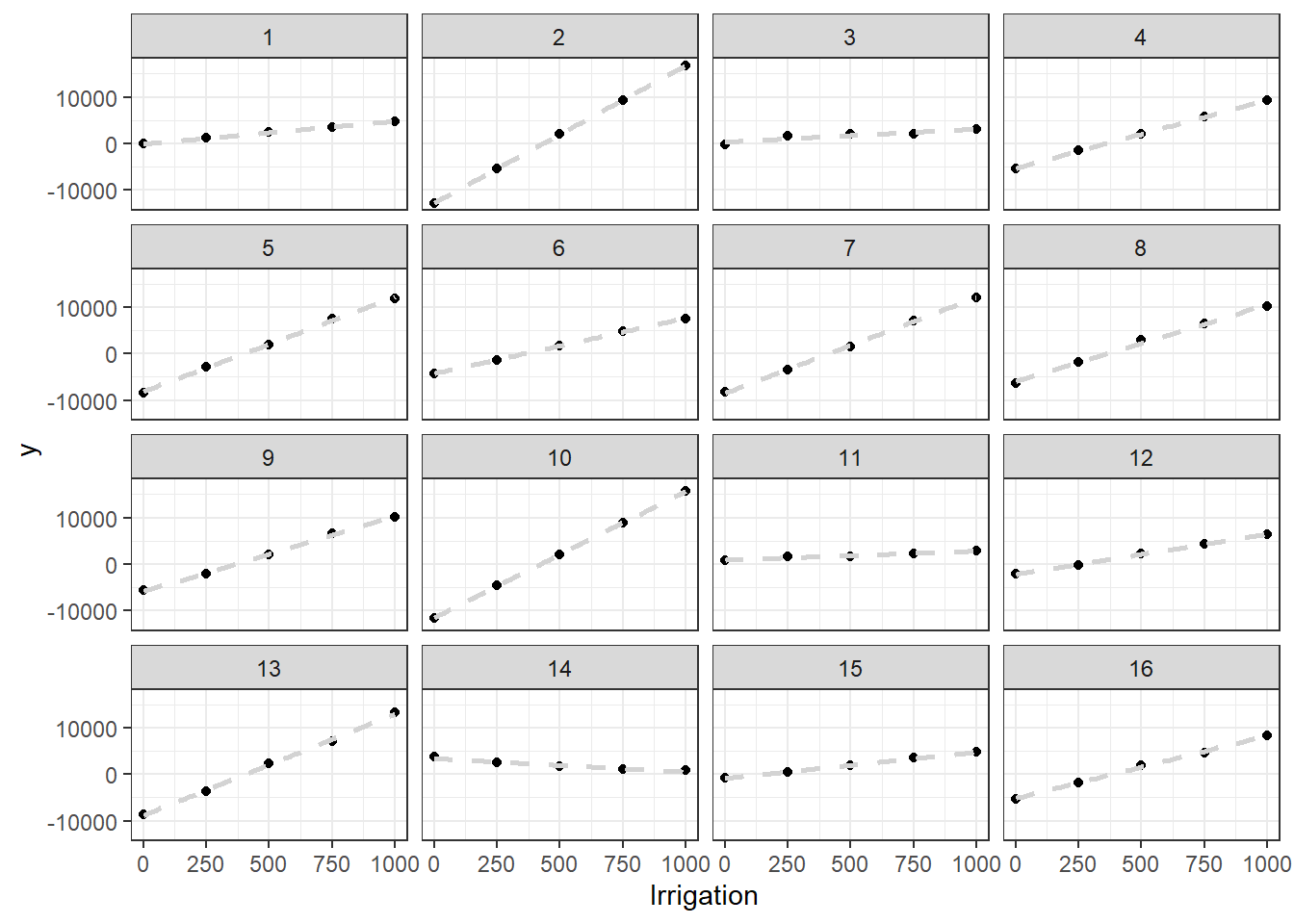
6.4 A more complex example: Effect of P nutrition on yield across farms
Read in the data:
pke.dat <- read.csv("data/raw/nigeria.pke.dat.csv")Create new variables to encode P fertilization treatment vs control (ignoring the other nutrients). First we create the special label encoding each treatment with a 0 or 1:
pke.dat$treat <- with(pke.dat,paste(i,p,k,e,om, sep = ""))Then we create a new variable that encodes the treatment as a factor and we order the data according to the different relevant factors:
pke.dat$treat.p <- NA
pke.dat$treat.p[pke.dat$treat=="00000"|pke.dat$treat=="10000"] <- "control"
pke.dat$treat.p[pke.dat$treat=="11000"|pke.dat$treat=="11100"|
pke.dat$treat=="11110"|pke.dat$treat=="11111"] <- "+p"
indices <- with(pke.dat, order(district, year, farm_id, treat.p))
pke.dat <- pke.dat[indices,]We also need to create a dummy variable to encode the random slopes:
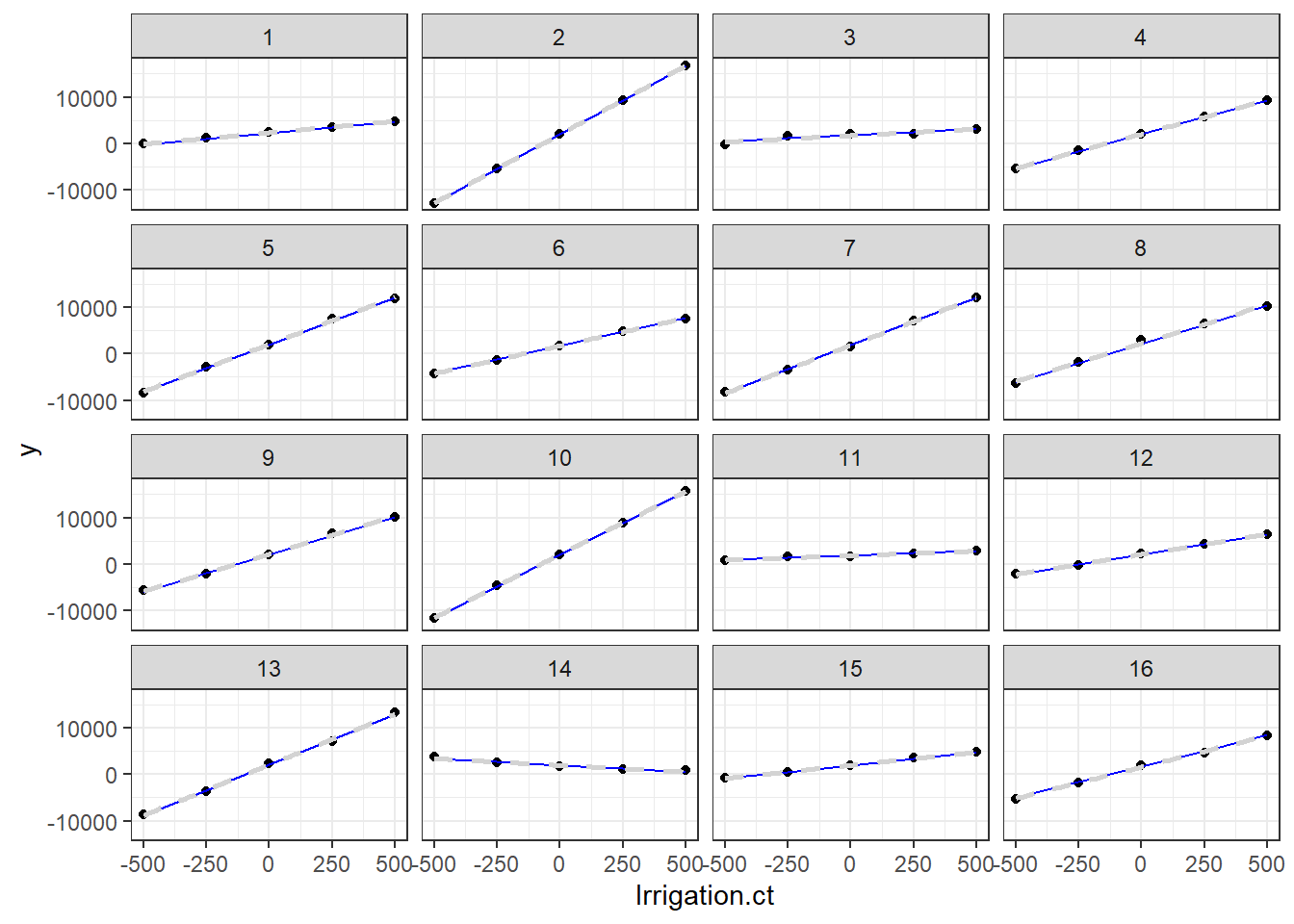
pke.dat$x1 <- ifelse(pke.dat$treat.p == "control", 0, 1)Exercise 6.3
- Fit two linear mixed models with random intercepts and slopes, one where these are correlated and one where they are not. Use
k,e,omandx1as fixed effects. and consider the following nested structure:district/year/farm_id. - Compare the models using the likelihood ratio test, which one is best?
- Continue with the best model and check the reliability of the parameter estimates using the profile plot.
- Check whether the assumption of normality and homoscedasticity of the residuals is met.
Solution 6.3
- We can fit the models as follows:
# with correlation
lmm1a <- lmer(estimated.grain.yield_kg_ha~k + e + om + x1 + (1 + x1| district / year /
farm_id), data= pke.dat)
# without correlation
lmm1b <- lmer(estimated.grain.yield_kg_ha~k + e + om + x1 + (1 + x1 || district / year /
farm_id), data= pke.dat)- We can compare the models using the likelihood ratio test:
anova(lmm1a, lmm1b)refitting model(s) with ML (instead of REML)Data: pke.dat
Models:
lmm1b: estimated.grain.yield_kg_ha ~ k + e + om + x1 + (1 + x1 || district/year/farm_id)
lmm1a: estimated.grain.yield_kg_ha ~ k + e + om + x1 + (1 + x1 | district/year/farm_id)
npar AIC BIC logLik -2*log(L) Chisq Df Pr(>Chisq)
lmm1b 12 11548 11604 -5762.2 11524
lmm1a 15 11551 11621 -5760.5 11521 3.5414 3 0.3154The best model is the one without correlation as adding correlation does not improve the likelihood well enough to justify the extra complexity.
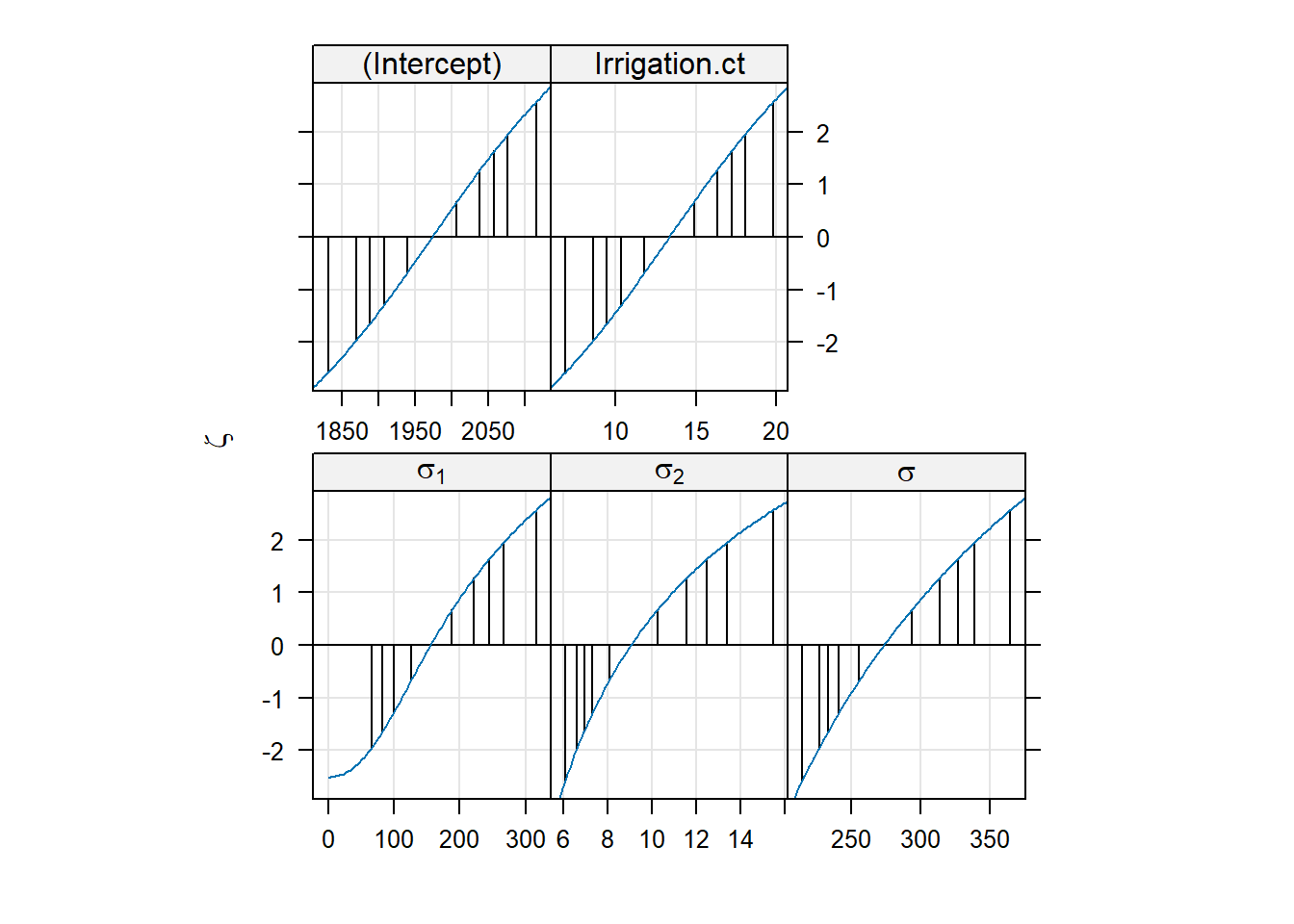
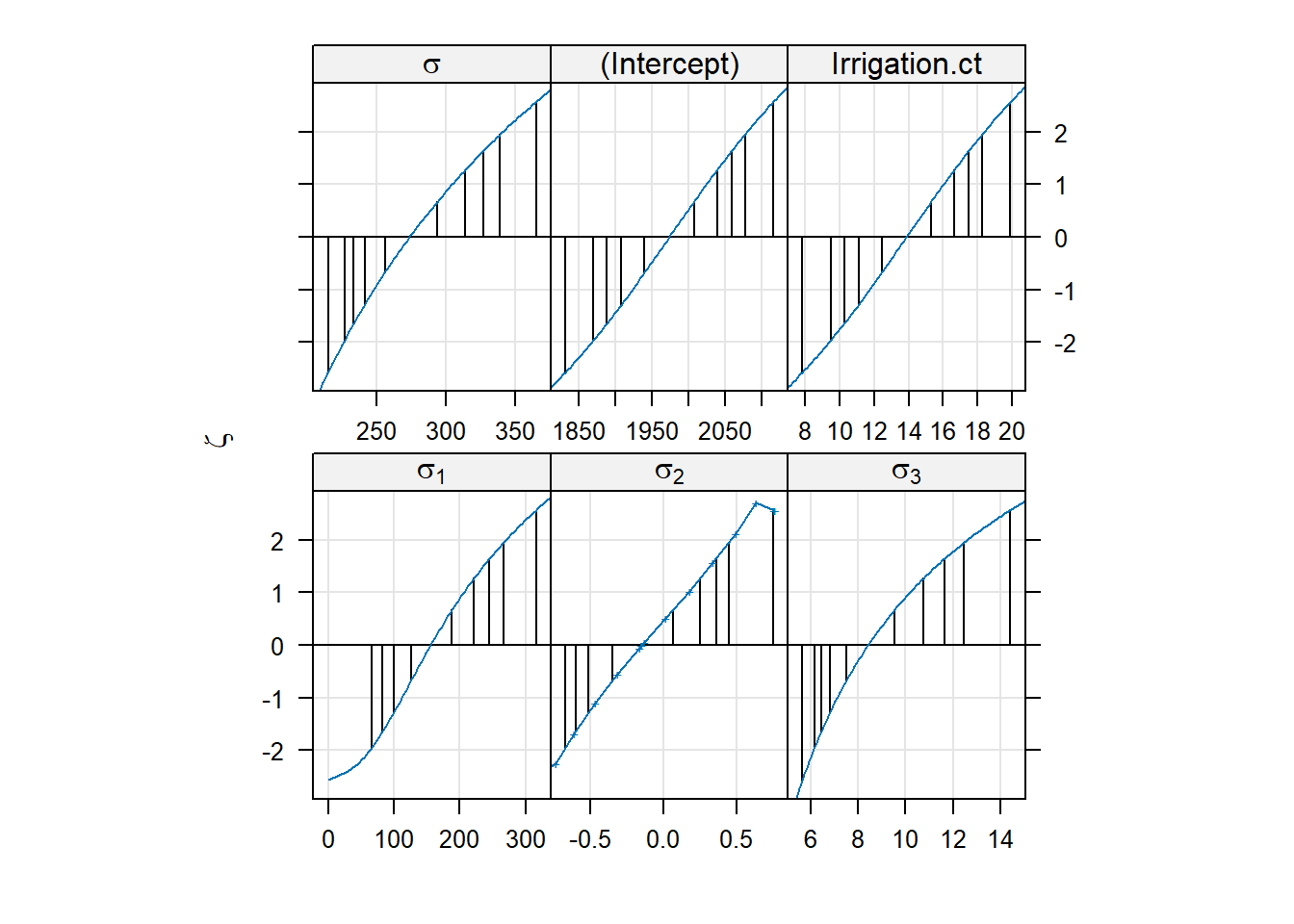
- We can check the reliability of the parameter estimates using the profile plot:
pr01 <- profile(lmm1b)Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
profile: using minstepWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
profile: using minstepWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
profile: using minstepWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
profile: using minstepWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
profile: using minstepWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
profile: using minstepWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
profile: using minstepWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
profile: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): unexpected decrease in
profile: using minstepWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstep
Warning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in nextpar(mat, cc, i, delta, lowcut, upcut): Last two rows have
identical or NA .zeta values: using minstepWarning in zetafun(np, ns): slightly lower deviances (diff=-1.81899e-12)
detectedWarning in FUN(X[[i]], ...): non-monotonic profile for .sig05xyplot(pr01, aspect = 1.3)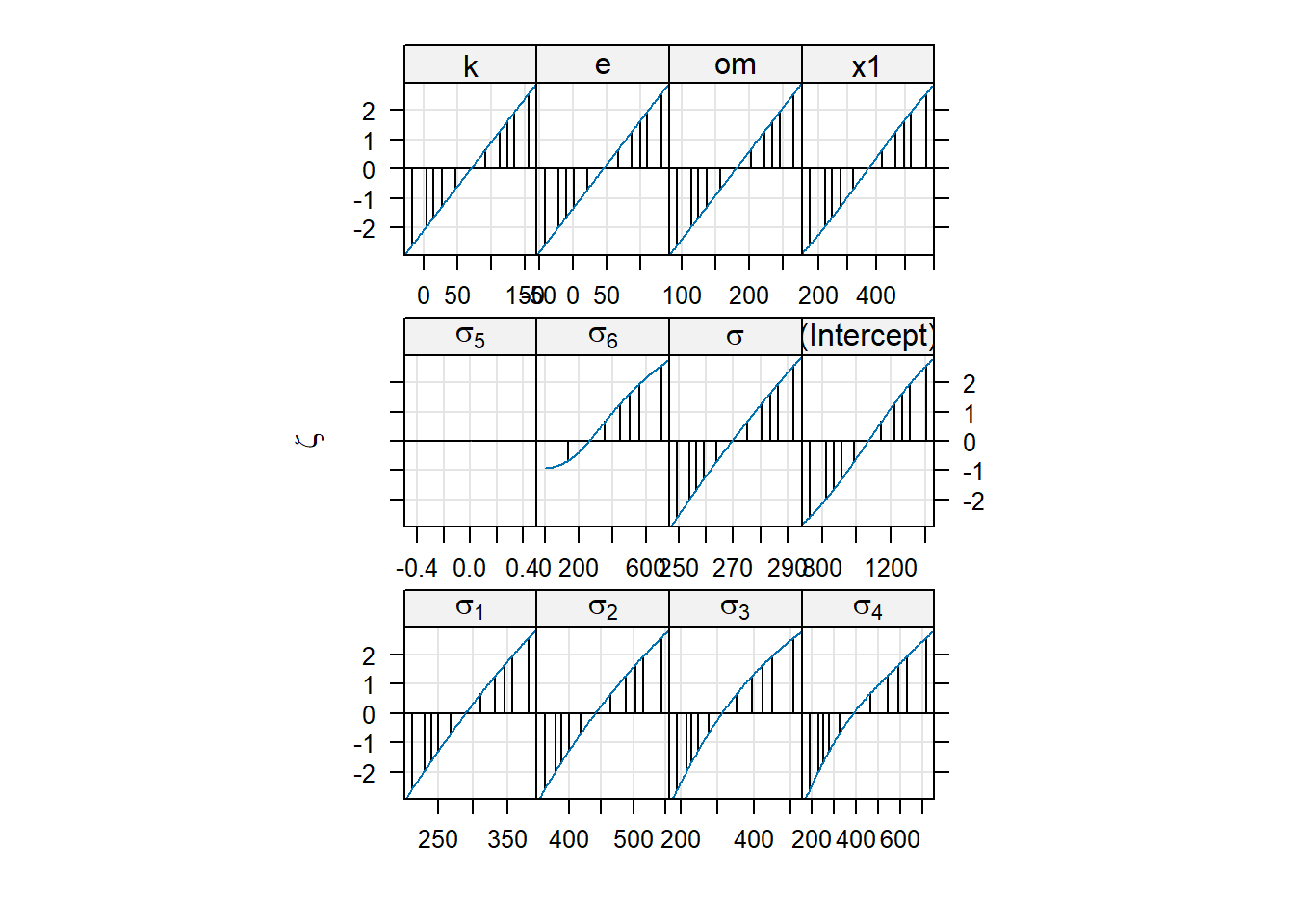
We can see that there are problems with the profiles of some of the variance components. We can use the summary to check which are those:
summary(lmm1b)Linear mixed model fit by REML. t-tests use Satterthwaite's method [
lmerModLmerTest]
Formula:
estimated.grain.yield_kg_ha ~ k + e + om + x1 + (1 + x1 || district/year/farm_id)
Data: pke.dat
REML criterion at convergence: 11476.8
Scaled residuals:
Min 1Q Median 3Q Max
-2.9298 -0.5216 -0.0490 0.4812 3.6955
Random effects:
Groups Name Variance Std.Dev.
farm_id.year.district x1 83251 288.5
farm_id.year.district.1 (Intercept) 194609 441.1
year.district x1 98602 314.0
year.district.1 (Intercept) 150370 387.8
district x1 5183 72.0
district.1 (Intercept) 82701 287.6
Residual 73039 270.3
Number of obs: 780, groups:
farm_id:year:district, 130; year:district, 26; district, 13
Fixed effects:
Estimate Std. Error df t value Pr(>|t|)
(Intercept) 1066.48 119.49 12.28 8.925 1.02e-06 ***
k 68.88 33.52 526.38 2.055 0.040382 *
e 44.09 33.52 526.38 1.315 0.188991
om 179.41 33.52 526.38 5.352 1.30e-07 ***
x1 371.01 76.64 11.51 4.841 0.000456 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Correlation of Fixed Effects:
(Intr) k e om
k 0.000
e 0.000 -0.500
om 0.000 0.000 -0.500
x1 -0.035 -0.219 0.000 0.000We can see that the estimates of the variance components for district are the ones where there seems to be some estimation problems. Note that this is the factor that sits on top of the hierarchy and therefore it is the one that has the most uncertainty associated with it. This is a common problem with nested random effects, where the top level of the hierarchy is the one that is most difficult to estimate.
- We can check the normality of the residuals using a QQ plot:
qqnorm(resid(lmm1b))
qqline(resid(lmm1b))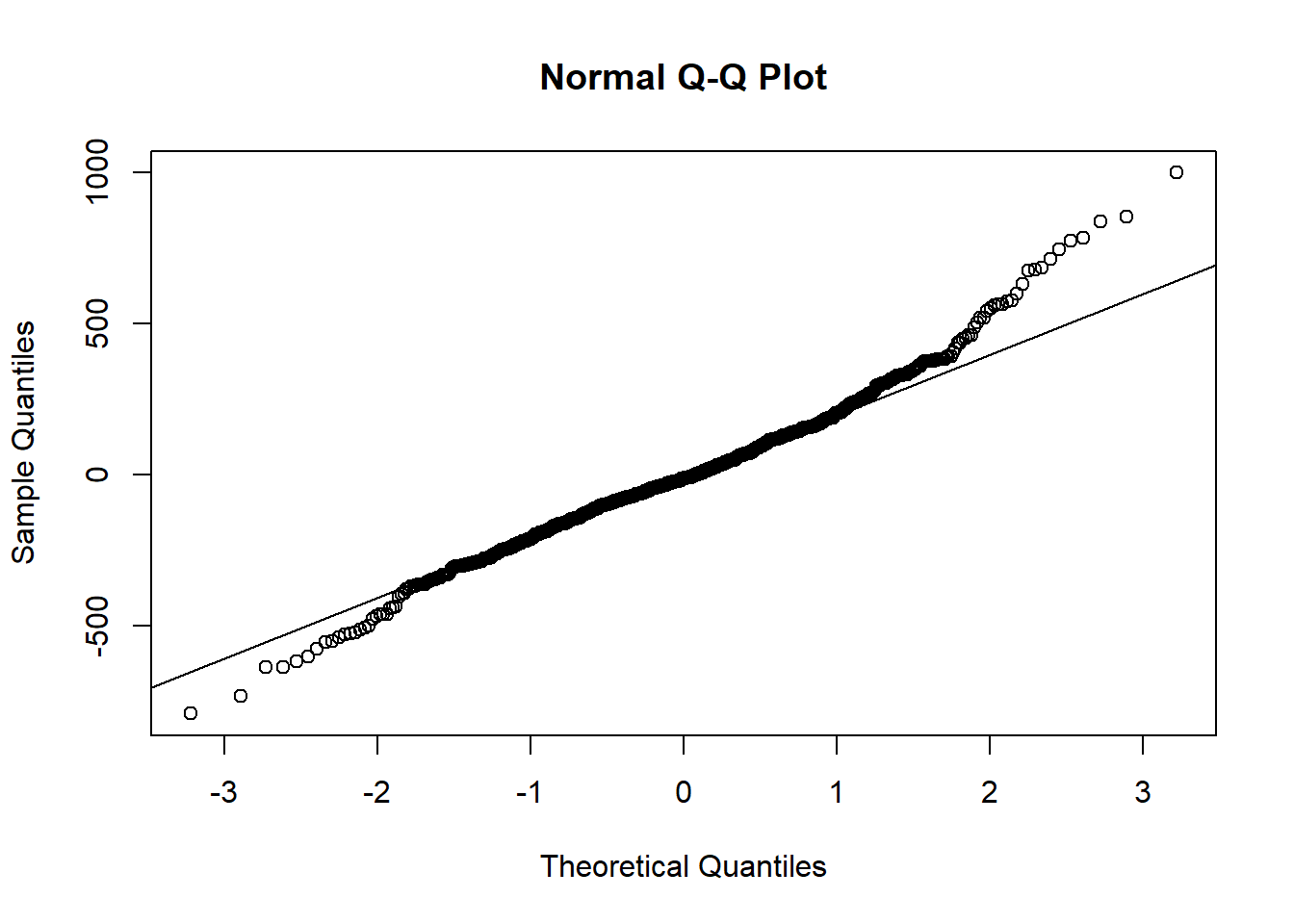
We see strong deviations from normality in the tails of the distribution. We can also use the plot function on the model object to check the homoscedasticity of the residuals:
plot(lmm1b)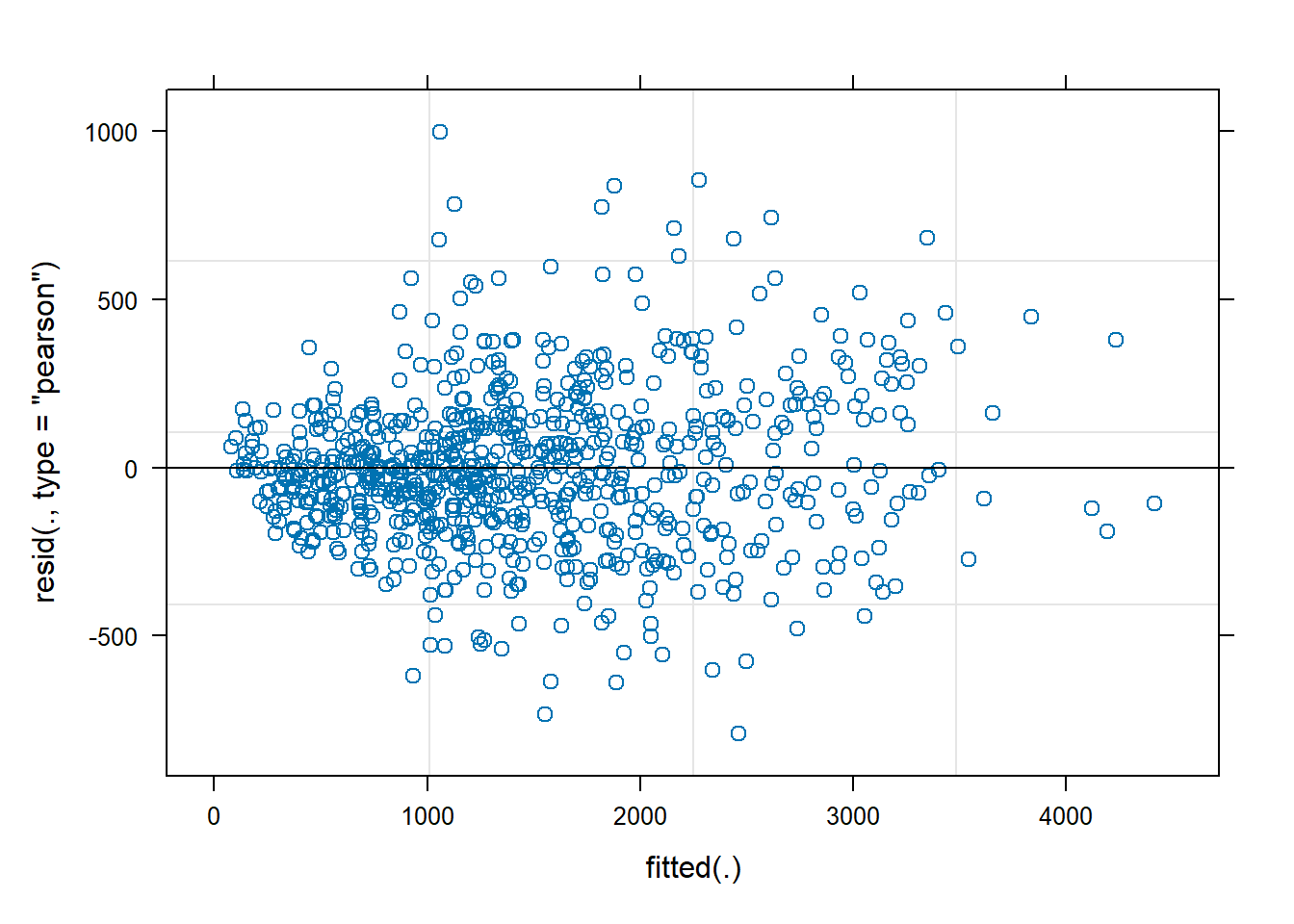
We can see that the residuals are not homoscedastic, there is a funnel effect where the variance is smaller at the lower end of the fitted values and larger at the higher end.
Let’s extract the variance components and show the contribution of each of these components to the total variance. First we extract the variance components and remove the correlation terms if present:
VC <- as.data.frame(VarCorr(lmm1b))
VC <- VC[is.na(VC$var2),]The sum of the variance components (including residuals) should match the variance of the data:
c(sum(VC$vcov), var(pke.dat$estimated.grain.yield_kg_ha))[1] 687755.8 752353.8We can see that is close enough (about 91% of the variance of the data). The mismatch is due to the fact that the variance components are estimated with some uncertainty, so the value sum(VC$vcov) actually has an uncertainty associated to it. Also, in general, estimates of variances tend to be biased downwards (the bias becomes smaller as the sample size increases).
We can visualize the contribution of each of the variance components to the total variance using a bar plot:
# make table with response variances
vc=VC$vcov
names(vc)<-paste(VC$grp,VC$var1) #set names to intercept/slope variance components
# make proportions
vc.prop<-vc/sum(vc)
# #order by (decreasing size)
vc.prop<-vc.prop[order(vc.prop, decreasing=T)]
# now plot
par_default <- par(no.readonly = TRUE) #save default parameters
par(mar=c(14,4,2,2)) #increase lower margin
barplot(vc.prop,las=2)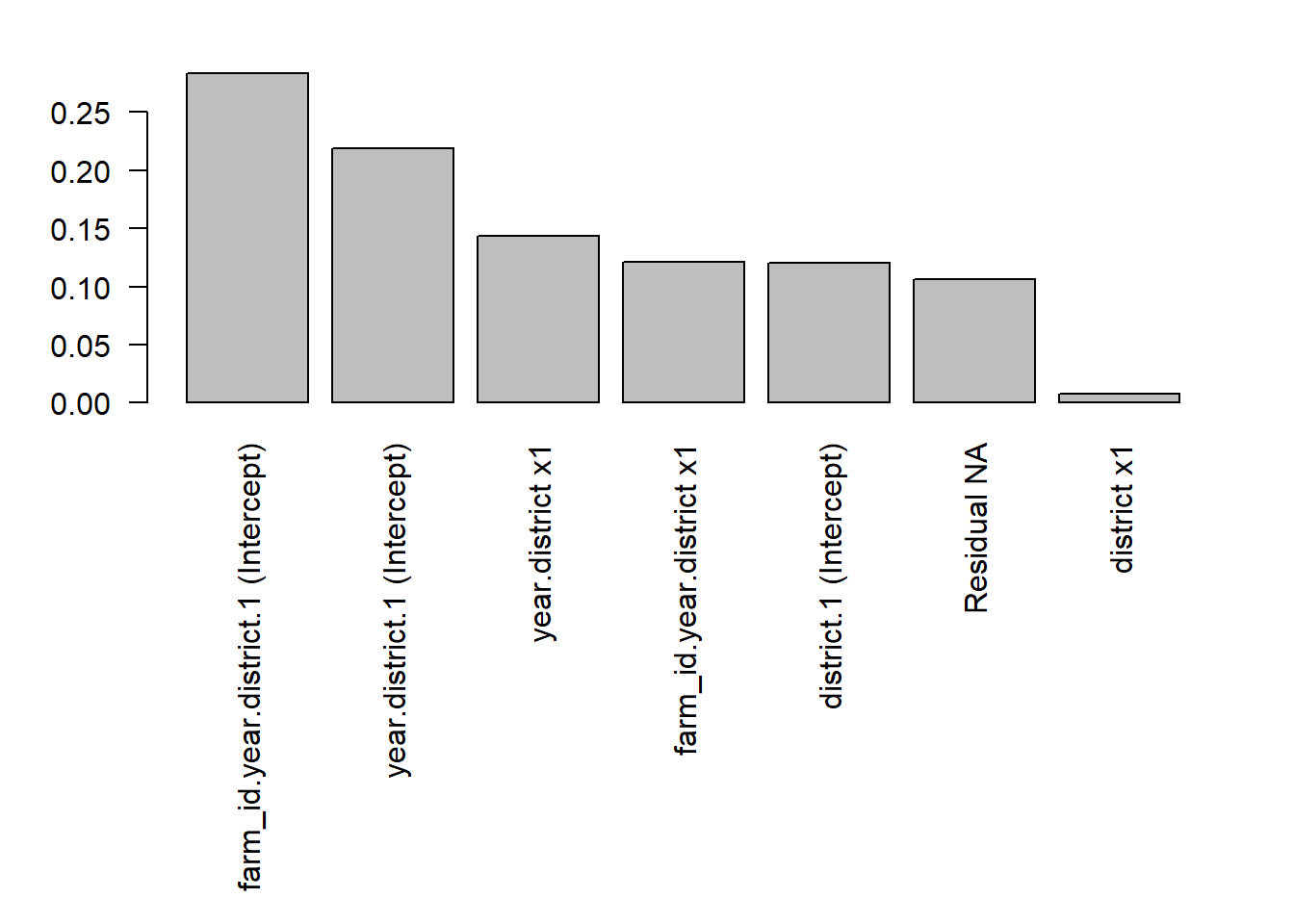
par(par_default) #reset to defaultWe can see that the farm and year components are the ones that contribute the most to the total variance, suggesting that there is substantial variation across farms and years in the effect of P fertilizer on yield.
To check on this, we can create subsets per district and year and fit separate models to compare the effects of P fertilizer. For example, let’s fit farm-level models to the data from Igabi and Kajuru districts in 2015:
dat.igabi.15 <- pke.dat[pke.dat$district == "igabi" & pke.dat$year == 2015,]
dat.kajuru.15 <- pke.dat[pke.dat$district == "kajuru" & pke.dat$year == 2015,]
lmm2.a<-lmer(estimated.grain.yield_kg_ha~k+e+om+x1+(1|farm_id),data=dat.igabi.15)
anova(lmm2.a)Type III Analysis of Variance Table with Satterthwaite's method
Sum Sq Mean Sq NumDF DenDF F value Pr(>F)
k 8109 8109 1 21 0.2110 0.6507
e 3071 3071 1 21 0.0799 0.7802
om 55042 55042 1 21 1.4323 0.2447
x1 20728 20728 1 21 0.5394 0.4708lmm2.b<-lmer(estimated.grain.yield_kg_ha~k+e+om+x1+(1|farm_id),data= dat.kajuru.15)
anova(lmm2.b)Type III Analysis of Variance Table with Satterthwaite's method
Sum Sq Mean Sq NumDF DenDF F value Pr(>F)
k 227571 227571 1 21 0.7212 0.405342
e 230157 230157 1 21 0.7294 0.402731
om 4034 4034 1 21 0.0128 0.911051
x1 3456966 3456966 1 21 10.9549 0.003333 **
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1We can see that in Kajuru the effect of P fertlizer on yield is significant, while in Igabi it is not.
Exercise 6.4
Extract the covariance matrix of the best model (
lmm1b) and visualize it. Explain the structure of the covariance matrix in relation to the data structureCompute the estimates of the coefficients and their standard errors using the linear algebra expressions we have taught you in the course (for fixed effects using the covariance matrix, the design matrix and the response variable).
Solution 6.4
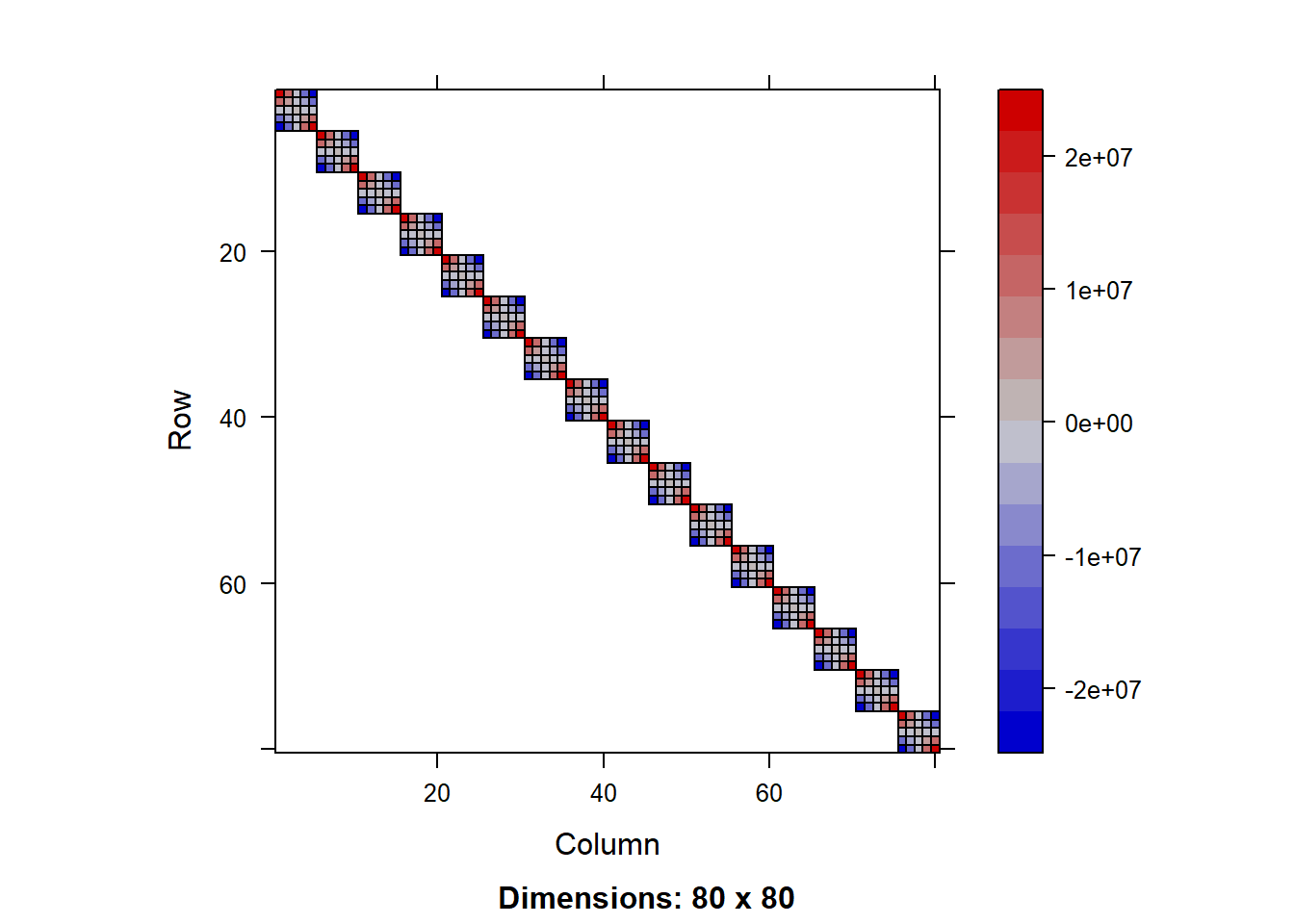
- We can extract the covariance matrix and visualize it as follows:
V = getV(lmm1b)
image(V[1:70,1:70])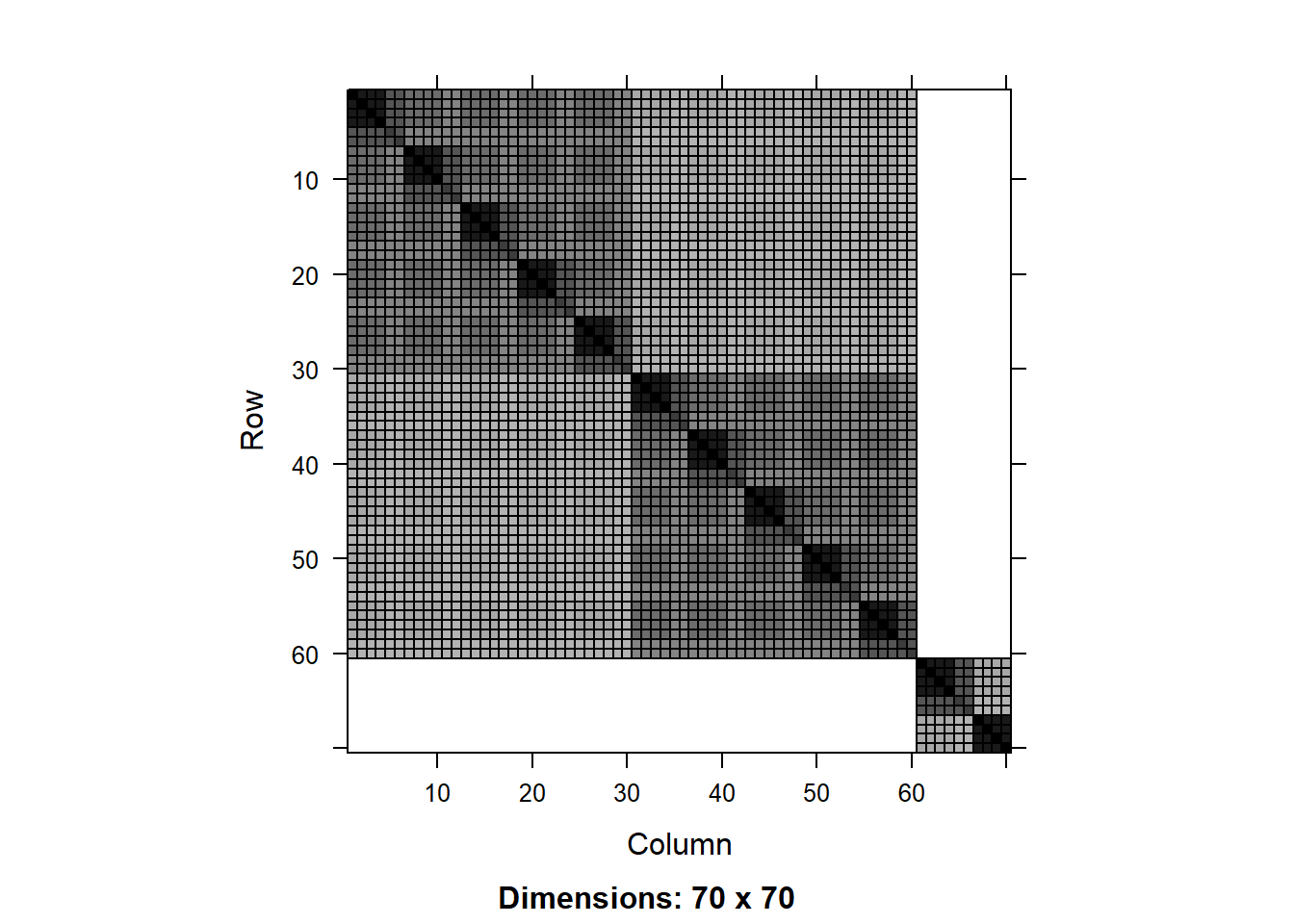
We can see a more complex structure of nested blocks of observations representing the different groups in the hierarchy. The big block of 60x60 represents the first district (bagwai, check table(pke.dat$district)), the two subblocks of 30x30 represent the two years (2015 and 2016), and the smaller blocks represent the farms.
- We extract the design matrix and the vector of response variables:
X <- model.matrix(lmm1b)
y <- pke.dat$estimated.grain.yield_kg_haWe apply the formulae to calculate the estimates of the coefficients and their standard errors:
# Estimates of coefficients
V_inv <- solve(V)
coef <- solve(t(X)%*%V_inv%*%X)%*%t(X)%*%V_inv%*%y
coef5 x 1 Matrix of class "dgeMatrix"
[,1]
(Intercept) 1066.47696
k 68.88258
e 44.08957
om 179.41296
x1 371.01233# Standard errors
COV <- solve(t(X)%*%V_inv%*%X)
SE <- sqrt(diag(COV))
SE(Intercept) k e om x1
119.48965 33.52122 33.52122 33.52122 76.63864 Compare to the output of the summary function:
summary(lmm1b)Linear mixed model fit by REML. t-tests use Satterthwaite's method [
lmerModLmerTest]
Formula:
estimated.grain.yield_kg_ha ~ k + e + om + x1 + (1 + x1 || district/year/farm_id)
Data: pke.dat
REML criterion at convergence: 11476.8
Scaled residuals:
Min 1Q Median 3Q Max
-2.9298 -0.5216 -0.0490 0.4812 3.6955
Random effects:
Groups Name Variance Std.Dev.
farm_id.year.district x1 83251 288.5
farm_id.year.district.1 (Intercept) 194609 441.1
year.district x1 98602 314.0
year.district.1 (Intercept) 150370 387.8
district x1 5183 72.0
district.1 (Intercept) 82701 287.6
Residual 73039 270.3
Number of obs: 780, groups:
farm_id:year:district, 130; year:district, 26; district, 13
Fixed effects:
Estimate Std. Error df t value Pr(>|t|)
(Intercept) 1066.48 119.49 12.28 8.925 1.02e-06 ***
k 68.88 33.52 526.38 2.055 0.040382 *
e 44.09 33.52 526.38 1.315 0.188991
om 179.41 33.52 526.38 5.352 1.30e-07 ***
x1 371.01 76.64 11.51 4.841 0.000456 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Correlation of Fixed Effects:
(Intr) k e om
k 0.000
e 0.000 -0.500
om 0.000 0.000 -0.500
x1 -0.035 -0.219 0.000 0.000